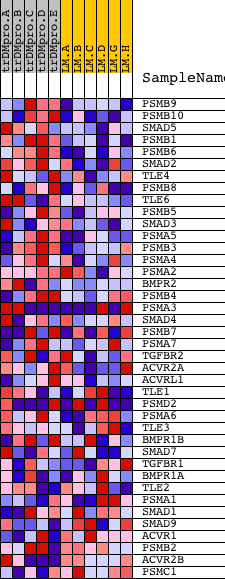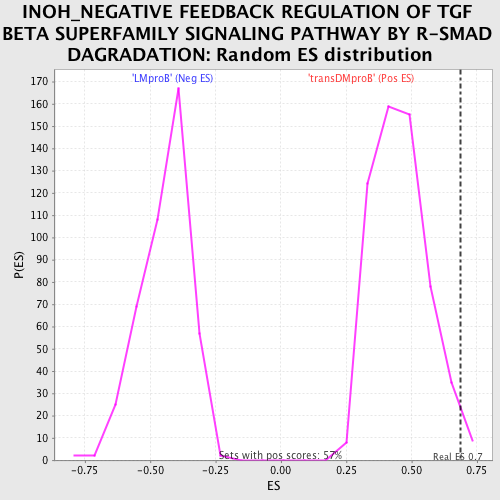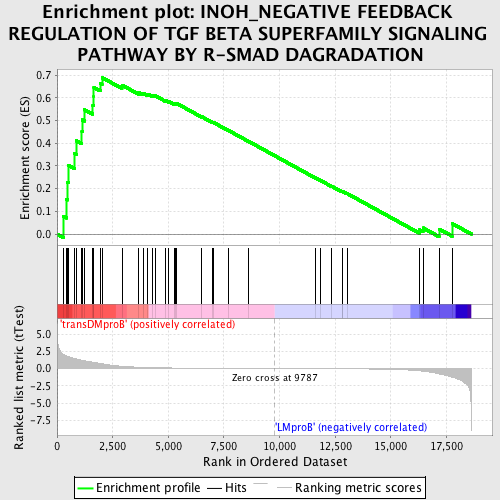
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_04_transDMproB_versus_LMproB.phenotype_transDMproB_versus_LMproB.cls #transDMproB_versus_LMproB |
| Phenotype | phenotype_transDMproB_versus_LMproB.cls#transDMproB_versus_LMproB |
| Upregulated in class | transDMproB |
| GeneSet | INOH_NEGATIVE FEEDBACK REGULATION OF TGF BETA SUPERFAMILY SIGNALING PATHWAY BY R-SMAD DAGRADATION |
| Enrichment Score (ES) | 0.6889555 |
| Normalized Enrichment Score (NES) | 1.5163006 |
| Nominal p-value | 0.01584507 |
| FDR q-value | 0.4301256 |
| FWER p-Value | 0.992 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | PSMB9 | 23021 | 277 | 2.071 | 0.0780 | Yes | ||
| 2 | PSMB10 | 5299 18761 | 416 | 1.833 | 0.1527 | Yes | ||
| 3 | SMAD5 | 21621 | 469 | 1.756 | 0.2287 | Yes | ||
| 4 | PSMB1 | 23118 | 508 | 1.706 | 0.3032 | Yes | ||
| 5 | PSMB6 | 9634 | 775 | 1.454 | 0.3541 | Yes | ||
| 6 | SMAD2 | 23511 | 864 | 1.376 | 0.4111 | Yes | ||
| 7 | TLE4 | 23720 3697 | 1115 | 1.187 | 0.4508 | Yes | ||
| 8 | PSMB8 | 5000 | 1132 | 1.179 | 0.5029 | Yes | ||
| 9 | TLE6 | 19669 | 1233 | 1.110 | 0.5472 | Yes | ||
| 10 | PSMB5 | 9633 | 1607 | 0.915 | 0.5682 | Yes | ||
| 11 | SMAD3 | 19084 | 1649 | 0.891 | 0.6060 | Yes | ||
| 12 | PSMA5 | 6464 | 1654 | 0.889 | 0.6456 | Yes | ||
| 13 | PSMB3 | 11180 | 1944 | 0.724 | 0.6625 | Yes | ||
| 14 | PSMA4 | 11179 | 2022 | 0.681 | 0.6890 | Yes | ||
| 15 | PSMA2 | 9631 | 2917 | 0.334 | 0.6558 | No | ||
| 16 | BMPR2 | 14241 4081 | 3678 | 0.190 | 0.6234 | No | ||
| 17 | PSMB4 | 15252 | 3866 | 0.166 | 0.6208 | No | ||
| 18 | PSMA3 | 9632 5298 | 4081 | 0.144 | 0.6158 | No | ||
| 19 | SMAD4 | 5058 | 4275 | 0.126 | 0.6110 | No | ||
| 20 | PSMB7 | 2795 14598 | 4408 | 0.117 | 0.6091 | No | ||
| 21 | PSMA7 | 14318 | 4864 | 0.092 | 0.5888 | No | ||
| 22 | TGFBR2 | 2994 3001 10167 | 5015 | 0.085 | 0.5845 | No | ||
| 23 | ACVR2A | 8542 | 5278 | 0.075 | 0.5737 | No | ||
| 24 | ACVRL1 | 22354 | 5322 | 0.074 | 0.5747 | No | ||
| 25 | TLE1 | 5765 10185 15859 | 5375 | 0.072 | 0.5752 | No | ||
| 26 | PSMD2 | 10137 5724 | 6511 | 0.043 | 0.5160 | No | ||
| 27 | PSMA6 | 21270 | 6976 | 0.035 | 0.4926 | No | ||
| 28 | TLE3 | 5767 19416 | 7035 | 0.034 | 0.4910 | No | ||
| 29 | BMPR1B | 8661 15152 4451 | 7723 | 0.023 | 0.4550 | No | ||
| 30 | SMAD7 | 23513 | 8609 | 0.013 | 0.4080 | No | ||
| 31 | TGFBR1 | 16213 10165 5745 | 11615 | -0.020 | 0.2471 | No | ||
| 32 | BMPR1A | 8660 4450 | 11852 | -0.023 | 0.2355 | No | ||
| 33 | TLE2 | 3378 | 12342 | -0.030 | 0.2105 | No | ||
| 34 | PSMA1 | 1627 17669 | 12805 | -0.037 | 0.1873 | No | ||
| 35 | SMAD1 | 18831 922 | 12826 | -0.038 | 0.1879 | No | ||
| 36 | SMAD9 | 15591 | 13032 | -0.042 | 0.1787 | No | ||
| 37 | ACVR1 | 4334 | 16280 | -0.327 | 0.0186 | No | ||
| 38 | PSMB2 | 2324 16078 | 16450 | -0.389 | 0.0270 | No | ||
| 39 | ACVR2B | 19276 | 17171 | -0.758 | 0.0222 | No | ||
| 40 | PSMC1 | 9635 | 17755 | -1.238 | 0.0463 | No |